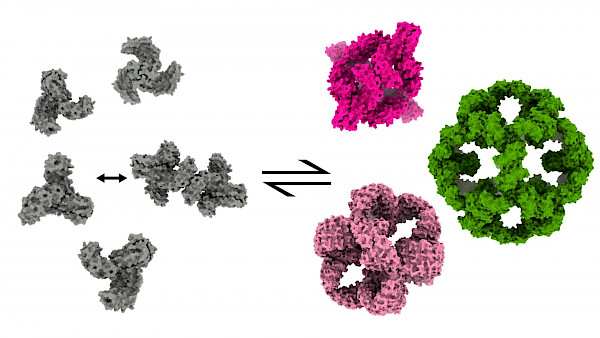
Research

Protein self-assembly is a ubiquitous phenomenon across all domains of life as well as viruses: protein building blocks are programmed to interact with one-another through supramolecular forces, adopting a wide variety of architectures ranging from unbounded crystals and filaments to bounded polyhedral assemblies.
The group of Protein Design and Self-Assembly focuses on dissecting the physical principles of protein self-assembly. While conceptually simple, the phenomenon of self-assembly entails a fine equilibrium of a number of physical properties, which determine the dynamics and structure of the assembly architecture.
Our group combines computational de novo protein design with protein production, in vitro model substrates and biophysical methods to systematically investigate the interplay between different types of interactions in the assembly process of protein-based materials.
The current first principle-based (Rosetta software suite) and AI-based methods, allows us to design rigid polyhedral protein assemblies with atomic-level accuracy that obey strict symmetry rules. We aim to expand the design process to dynamic and responsive protein assemblies, akin to natural assemblies, by exploring protein structural flexibility and by coupling assembly to biologically relevant processes, e.g. cargo encapsulation and membrane envelopment.